ML in Musculoskeletal Imaging

Hamstring Muscles Tear
Niamh Belton
Principal Investigators: Dr Kathleen Curran(UCD) and Dr Aonghus Lawlor(UCD)
Noisy data present in medical imaging datasets can often aid the development of robust models that are equipped to handle real-world data. However, if the bad data contains insufficient anatomical information, it can have a severe negative effect on the model's performance. We propose a novel methodology using a semi-supervised Siamese network to identify bad data. This method requires only a small pool of 'reference' medical images to be reviewed by a non-expert human to ensure the major anatomical structures are present in the Field of View. The model trains on this reference set and identifies bad data by using the Siamese network to compute the distance between the reference set and all other medical images in the dataset. This methodology achieves an Area Under the Curve (AUC) of 0.989 for identifying bad data. Code will be available at https://git.io/JYFuV.
Knee Injuries
Niamh Belton, Adil Dahlan, Misgina Tsighe Hagos and Ronan Hearne
Supervisors: Dr Kathleen Curran (UCD), Dr Aonghus Lawlor(UCD) and Assoc Professor Brian Mac Namee(UCD)
Knee injuries are one of the most common sporting injuries. Such injuries often include Anterior Cruciate Ligament (ACL) tears and meniscus tears. These injuries have been known to be detrimental to athletes careers. Early diagnosis of knee injuries is crucially important in order to intervene early with appropriate rehabilitation. By harnessing data from Stanford University Medical Centre, first published by Bien et al. (2018), we can build highly accurate models to automatically detect and diagnose knee injuries. Before modelling, we perform data augmentation techniques such as flipping the image and rotating it. We achieved comparable results to the current state of the art by employing the modern CNN architecture ResNet18. We also ensure our solution is interpretable by visualizing the images with Grad-Cam. This initial exploration gives encouraging results to do further modelling


Hamstring Muscles Tear
Adil Dahlan, Dr Nicholas McCarthy and Brendan StJohn
Principal Investigators: Dr Kathleen Curran(UCD) and Dr Aonghus Lawlor(UCD)
Muscle injuries are common in sport and account for substantial time lost from training and competition. The hamstrings are the most frequently injured muscle group and hamstring muscle injuries alone result in an average of 90 days missed per club per season in professional soccer. Muscle injuries are also common in rugby union, Australian Rules football, basketball and other Olympic sports. In collaboration with clinicians and sports club we are building a classifier for hamstring muscle tears.
Zebrafish Vertebrae Segmentation and Quantifying Spine Curvature
Adil Dahlan, Katie Noonan and Yushi Yang
Supervisors: Dr Kathleen Curran (UCD), Dr John Healy (UCD) and Dr Erika Kague (University of Bristol)
A lot has been discovered about the genetics of monogenetic bone diseases, however it is only recently that the elaborated genetic architecture behind complex conditions such as osteoporosis, osteoarthritis, scoliosis and disc degeneration has been revealed. Zebrafish are bony fresh water model organisms that have been used to investigate the genetics and molecular mechanisms of skeletal conditions. Due to the easy genetic manipulation in zebrafish and over 70% genetic similarity with humans, it is possible to validate and test how identified genes are involved in different skeletal conditions accelerating therapeutic discoveries in the bone field. We are currently developing a neural network that could automatically segment the zebrafish vertebral column and quantify scoliosis in order to systematically identify the effect of each mutant gene on the bones architecture and spine structure.

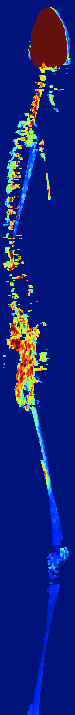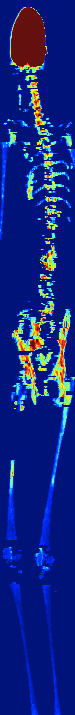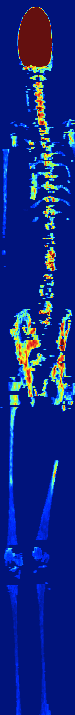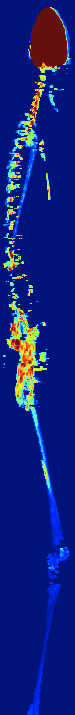
AUTOMATIC BONE MARROW SEGMENTATION FOR PET/CT IMAGING
IN MULTIPLE MYELOMA
Dr Patrick Leydon
Supervisor: Dr Kathleen Curran (UCD), Dr Derek Greene (UCD) and Dr. Martin O'Connell (UCD)
Multiple myeloma (MM) is a malignant hematologic disorder characterized by bone marrow infiltration with neoplastic
plasma cells. Approximately 10% of all hematologic cancers are related to MM. Whole-body 18F-FDG PETCT is an extremely useful imaging tool for the assessment of patients with MM.
We developed a novel approach to automatically segments bone
marrow regions of interest on both the PET and CT datasets.
Synthetic PET scans Using C-GANs for Healthy Bone Marrow Baseline Image Generation
Dr Patrick Leydon
Supervisor: Dr Kathleen Curran (UCD), Dr Derek Greene (UCD) and Dr. Martin O'Connell (UCD)
A Conditional-Generative Adversarial Network (C-GAN) has been used for a supervised image-to-image translation task which outputs a synthetic PET scan based on real patient CT data. The network is trained using only data of patients with healthy bone marrow metabolism. This allows for a patient specific synthetic healthy baseline scan to be produced. This can be used by a clinician for comparison to real PET data in the absence of a baseline scan or to aid in the diagnosis of conditions such as Multiple Myeloma which manifest as changes in bone marrow metabolism.
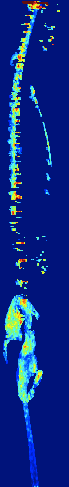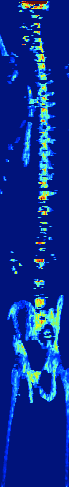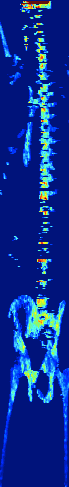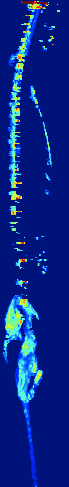

Probabilistic Lenke Classification (PLC)
Darragh Maguire
Supervisors: Dr Kathleen Curran(UCD and Dr John Healy(UCD)
Clinical Collaborators: Mr Michael Dodds (Consultant Orthopaedic Surgeon), Mr Connor Green (Consultant Orthopaedic Surgeon), Prof Eoin Kavanagh (Consultant Radiologist), Dr Rosanne-Sara Lynham (Paediatric Orthopaedic Registrar) and Dr Ivan Welaratne (Specialist Registrar in Radiology)
The aim of this project was to develop novel computational methods to improve clinical assessment of scoliosis. All of the developed tools are free and open-source, with complete documentation and code available from GitHub.
PLC WebApp was developed in this study to enable orthopaedic surgeons to account for interobserver variability in Lenke classification. Each angle measured by a clinician is subject to an inherent bias. With PLC, a patient's true underlying Cobb angle is modelled as a normal distribution around the measured angle. This allows for calculation of the probability of each Lenke classification, providing an objective indicator of the level of uncertainty in classification for a given patient.
Use the WebApp or find the source code in this repository.
Website: https://www.scoliosistools.com/
Automatic Scoliosis Assessment
Darragh Maguire
Supervisors: Dr Kathleen Curran(UCD and Dr John Healy(UCD)
Clinical Collaborators: Mr Michael Dodds (Consultant Orthopaedic Surgeon), Mr Connor Green (Consultant Orthopaedic Surgeon), Prof Eoin Kavanagh (Consultant Radiologist), Dr Rosanne-Sara Lynham (Paediatric Orthopaedic Registrar) and Dr Ivan Welaratne (Specialist Registrar in Radiology)
Deep learning was applied to develop a system for automatic calculation of Cobb angles in spinal x-rays, aiming to improve the accuracy and repeatability of clinical measurements. Our proposed tool can locate each vertebra, define the vertebral endplates, determine the most sloped vertebrae, and calculate the PT, MT, and TL/L Cobb angles in a given x-ray.
See a sample of performance or find the source code in this repository.
website: https://www.scoliosistools.com/


